Cresset successfully achieves SOC 2 compliance
Cresset is pleased to announce the successful completion of its SOC 2® (System and Organization Controls 2) Type 1 audit, achieving ...
News
Molecular Dynamics (MD) in Flare demonstrates how your molecular system evolves in time. It permits the exploration of the configurational space of your molecular system, providing insight into conformational stability and atomistic mechanisms. The domain of applicability of MD includes soluble proteins and, within Flare, MD can be seamlessly applied to the study of membrane proteins, biological systems which are typically challenging to initialize and model.
In this article, we will showcase how effortlessly membrane protein MD simulations can be configured, executed, and analyzed within Flare.
The first key step to set-up a membrane protein system is that of obtaining a protein structure in the correct orientation and position with respect to a model membrane bilayer. Only then, the chosen bilayer can be built around the protein and the MD simulation run (Figure 1).
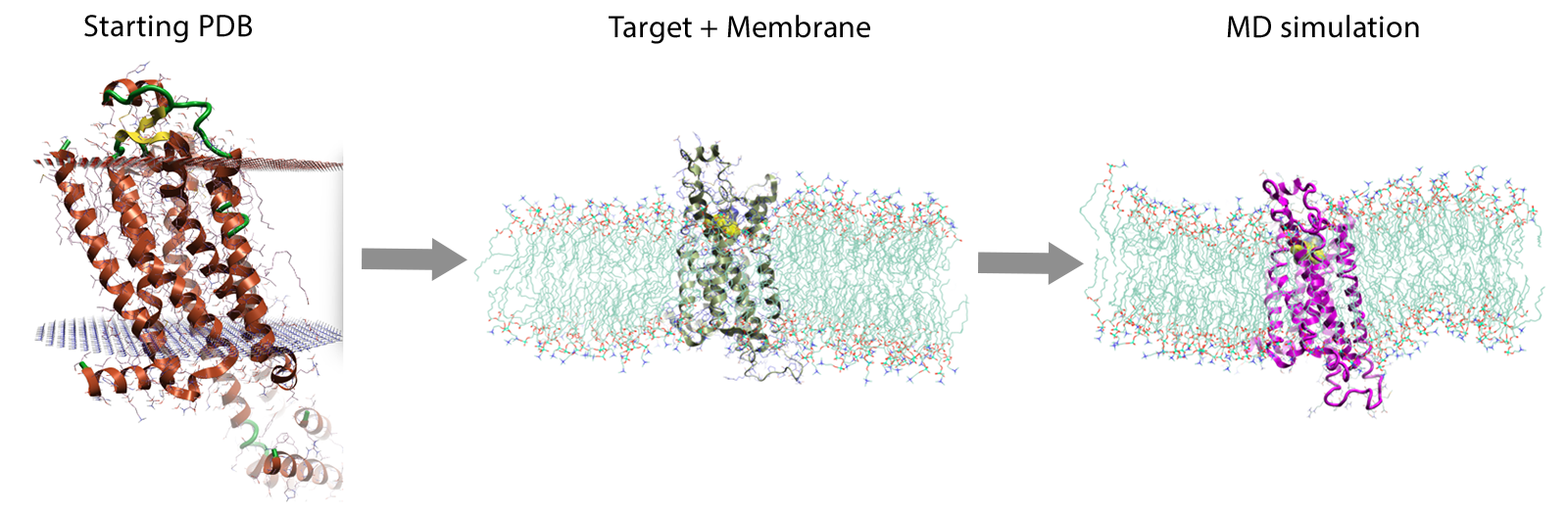
Figure 1. The workflow to set up membrane protein simulations in Flare
Proteins can be downloaded in the correct orientation with respect to the membrane from the Orientations of Proteins in Membranes (OPM) database directly through Flare’s PDB downloader. The URL path to the OPM database needs to be 'un-hashed' whereas the URL paths to the protein databank need to be 'hashed', forcing Flare to download the protein from the OPM database only (Figure 2). Once imported, Flare can prepare the protein structure, fixing any structural issues that will hinder running an MD simulation.
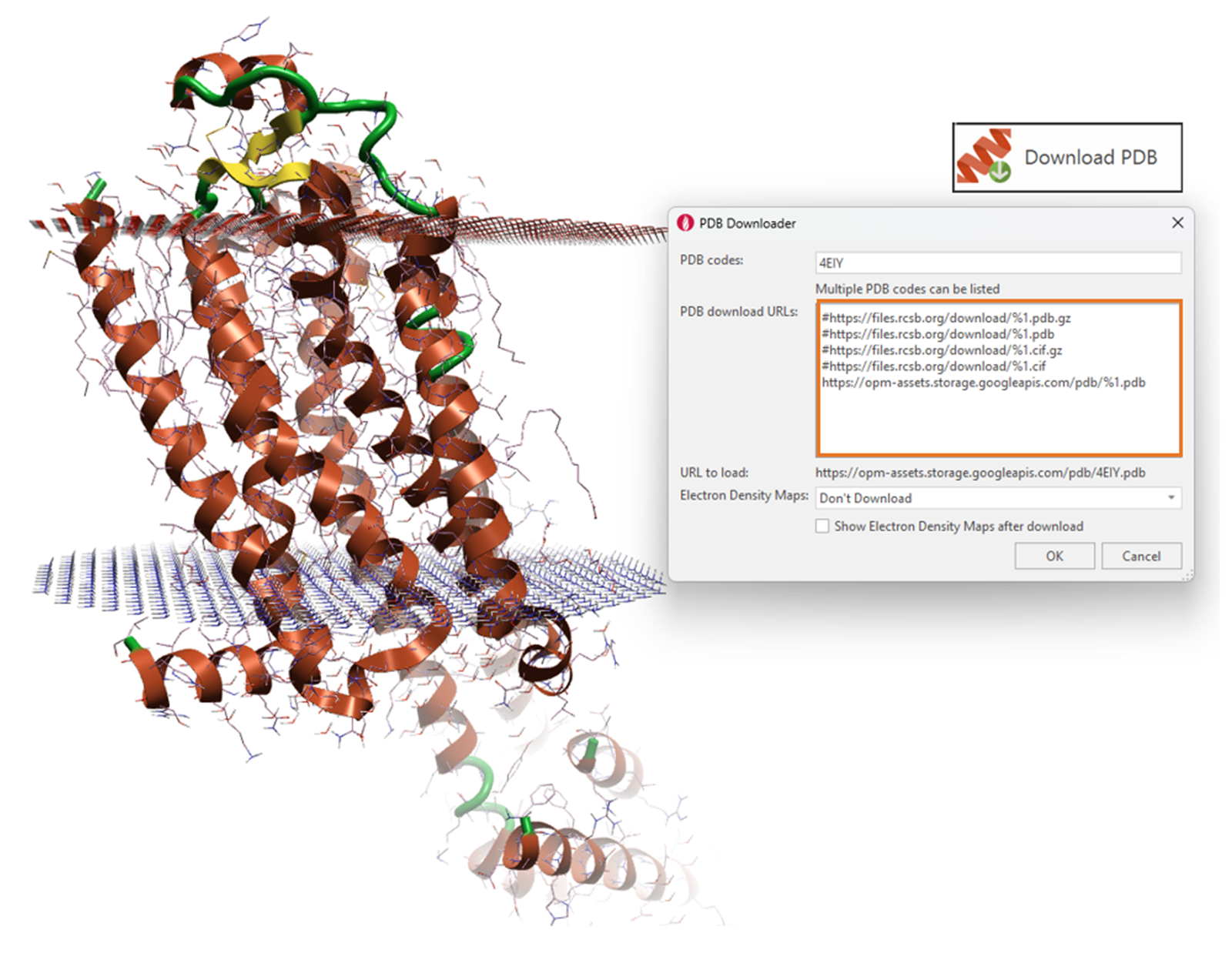
Figure 2. Downloading proteins in the correct orientation with respect to their parent membrane from the OPM database
By downloading the protein from the OPM database, the prepared protein structure is ensured to exist within the correct coordinate frame with respect to the membrane. Understandably, if your protein is not present in the OPM database, you can use Flare to download a homologous protein, from which you can sequence-align and superpose your structure onto. This will position your protein into the membrane.
Flare will build the lipid bilayer for your prepared membrane protein, within the process of setting up and running the MD simulation. Within the Dynamics calculation window, you will choose to run the calculation using OpenFF with membrane (Figure 2). This will apply the OpenFF 2.0 small molecule forcefield for the ligand, Amberff14sb for the protein and you can choose either a POPC or POPE lipid bilayer. In Figure 3, the vertical space that will be occupied by the lipid bilayer is shown as the blue cuboid.
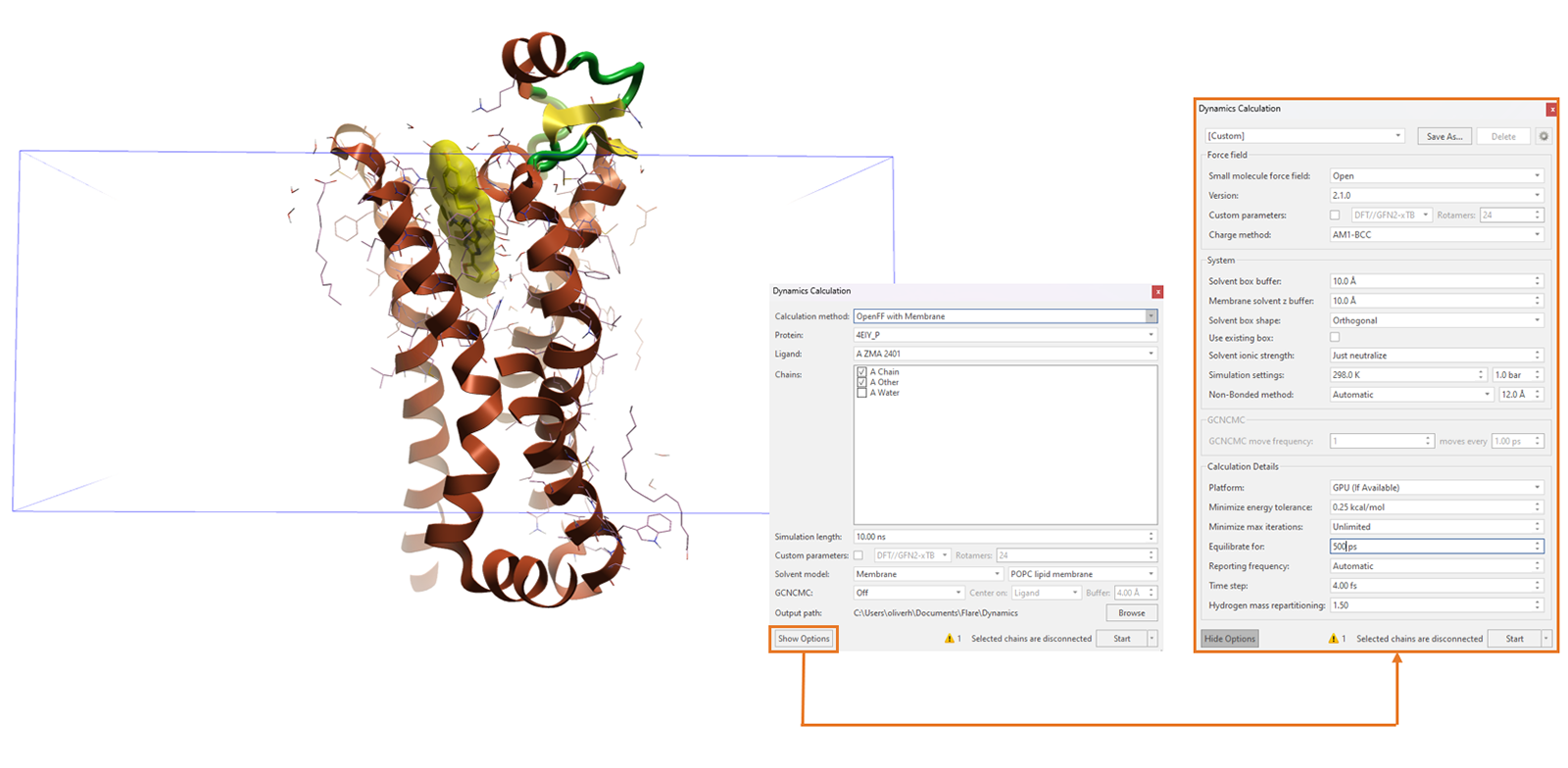
Figure 3. Building the lipid bilayer and running membrane protein MD simulations in one pre-configuration step.
As with any MD simulation, you have the option to lengthen or shorten the simulation time, compute custom ligand torsion parameters and include GCNCMC water sampling during the equilibration phase of your simulation. Furthermore, within the Show Options dialogue, you have the freedom to increase the equilibration time for your MD simulation, which may be necessary with very large protein-lipid bilayer systems that take longer to stabilize.
Flare offers a large suite of MD analysis tools, enabling the user to thoroughly analyse a simulation. Most notably, Flare can calculate protein RMSD, RMSF and radius of gyration as a function of time, providing insight into the protein's conformational stability as embedded within the lipid layer. These visual analysis tools help to interpret the protein function and behaviour, aiding understanding and enabling deeper insights when watching the final simulation and viewing the conformational dynamics in real time (Figure 4).
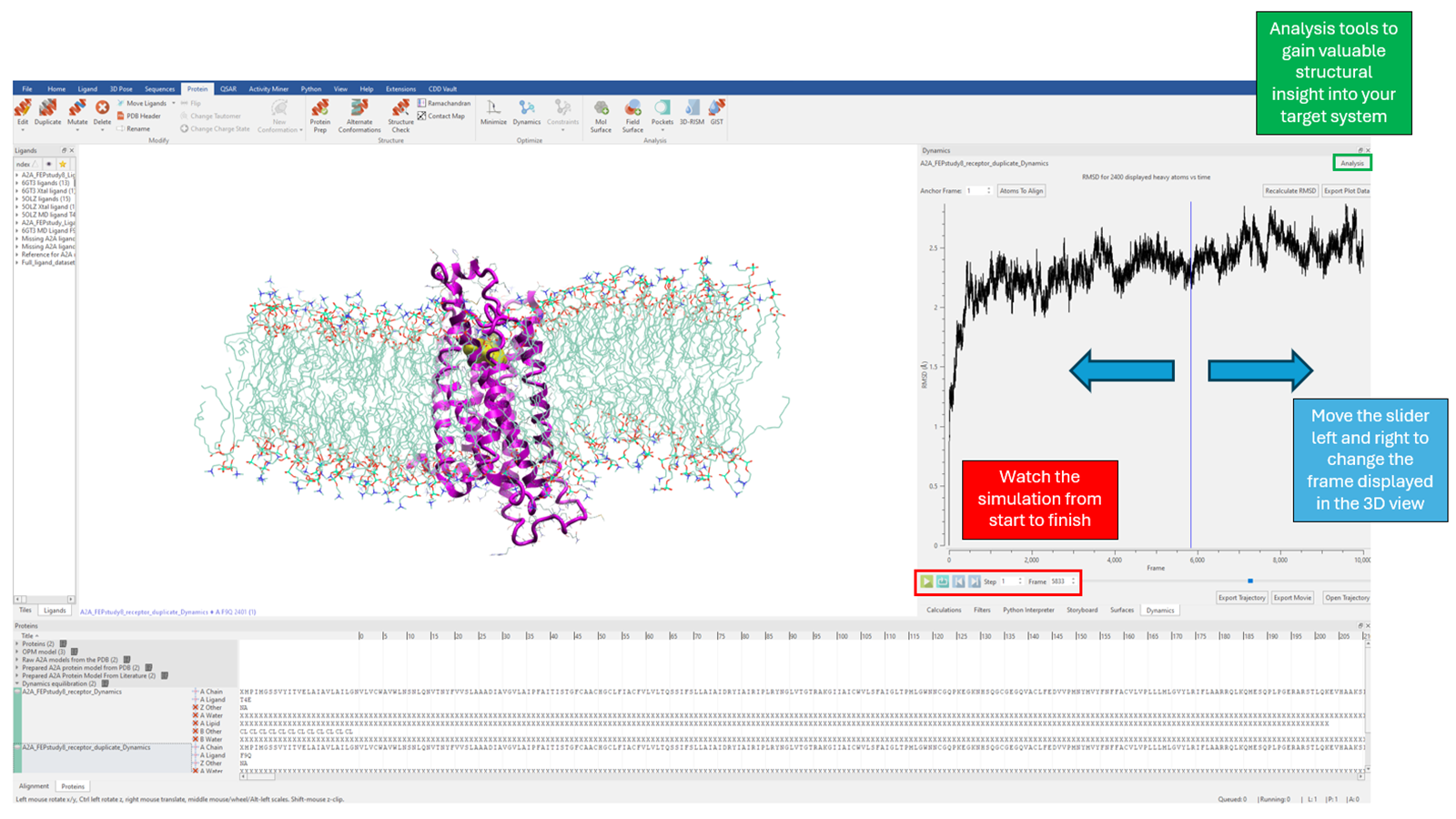
Figure 4. Viewing and structurally analyzing the conformational dynamics of your target system
The structural features of the lipid bilayer should remain stable over the course of the simulation. Particularly, the protein and lipid bilayer should favourably interact over the course of the simulation, not presenting any evidence of membrane collapse and demonstrating a conservation in the position of the membrane centre on the Z-axis (the axis perpendicular to the plane of the membrane). Notable symptoms of an unstable membrane protein simulation to watch out for include:
Stable contacts between the protein and lipid should keep the distance between them stable over time. If the lipid moves away from the protein over the course of the simulation, creating a large opening, this indicates that the protein has likely been inaccurately inserted into the membrane along the Z-axis. To test for this issue in Flare, one can change the display style of the lipid to CPK, orient the system along the Z-axis and re-watch the trajectory looking out for large openings created about the protein (Figure 5). When in doubt, one can track the distance between atoms using Flare's measurement option, specifically, tracking the distance between any two (user-defined) atoms over time.
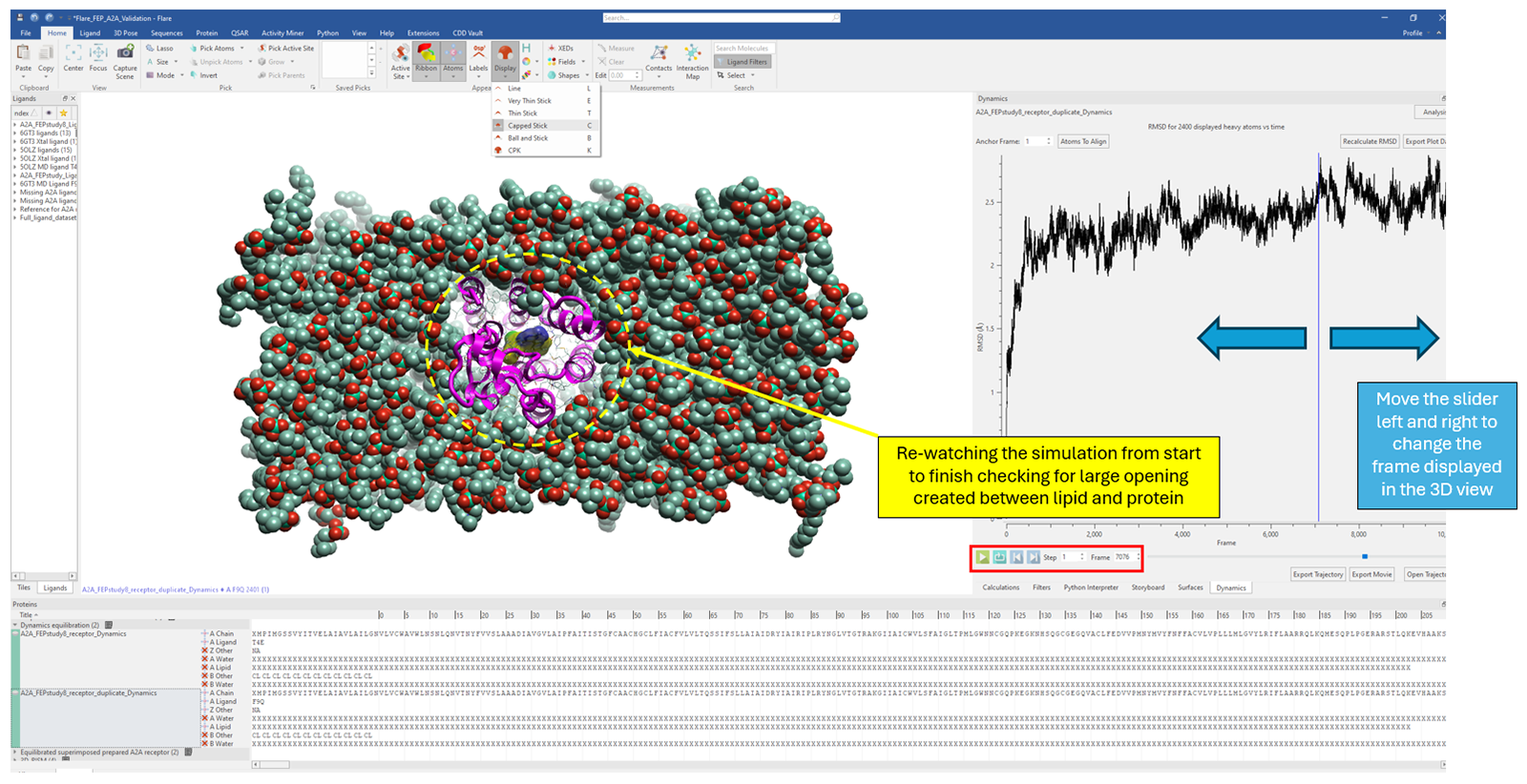
Figure 5: Changing the lipid display style to CPK and re-watching the simulation to inspect the distance between lipid and protein over time.
The hydrocarbon chains of the lipid bilayer are inherently flexible and should demonstrate this flexibility over the course of the simulation. Frozen hydrocarbon chains, which show no movement over the dynamical trajectory, are referred to as 'gelified'. This event typically arises as a result of poor forcefield parameterization. The effect of this, is that your membrane is non-physically rigid and therefore you lose confidence that you are sampling the true configurational space of the system. This can be easily elucidated through visual inspection of the membrane protein MD trajectory.
This refers to the observation that the lipid bilayer curves throughout the simulation, typically indicating that the simulation box is too small and/or the simulation pressure is too high (Figure 6). Analogous to the point above, this leads to a biophysical conformation that is not representative, lowering the accuracy and interpretability of your membrane protein simulation. Naturally, an extension to this is the observation of lipid bilayer envelope opening at the periodic boundaries. Opposite to Figure 6, this triggers the formation of a concave curvature at the centre of the Z-axis where the protein is embedded.
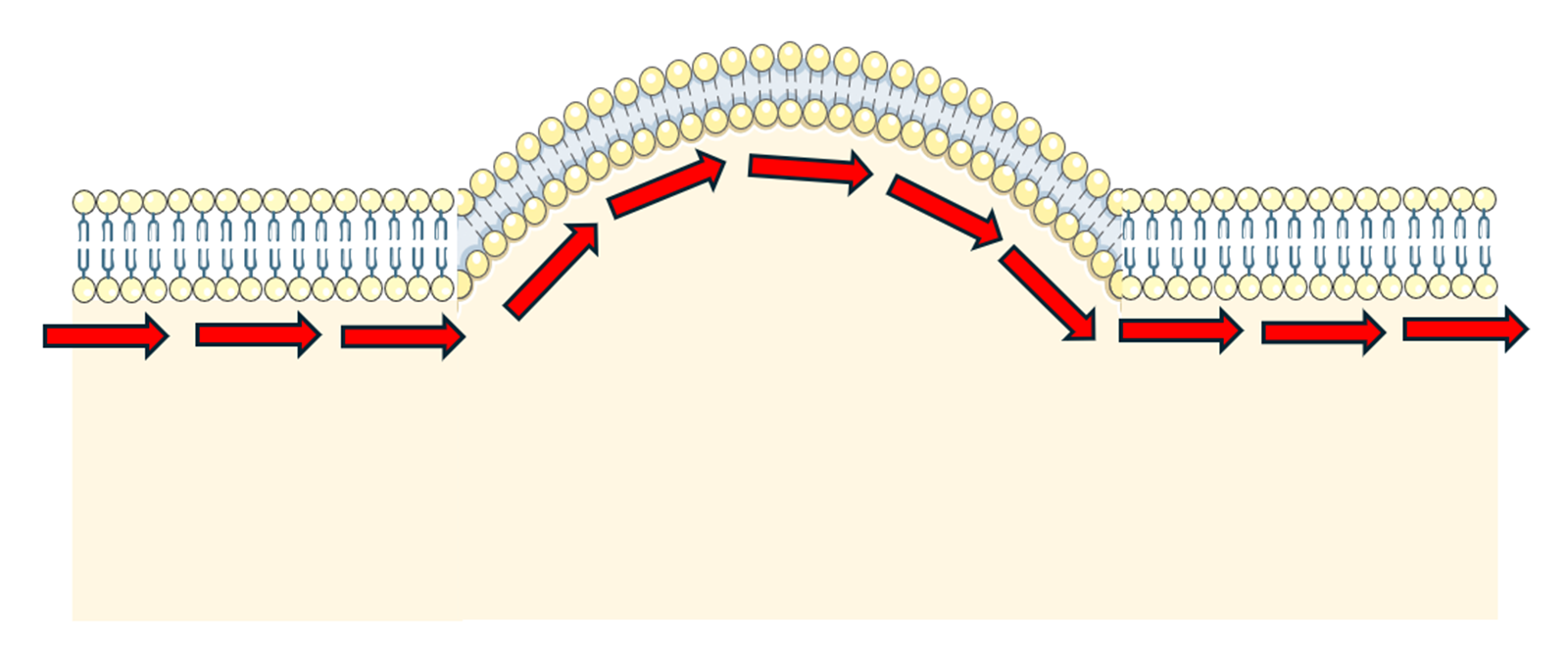
Figure 6. Undulation in the lipid bilayer. This figure was partly generated using Servier Medical Art, provided by Servier, licensed under a CC BY 4.0 license (CC BY 4.0 Deed | Attribution 4.0 International | Creative Commons).
In this article, we have discussed how membrane protein MD simulations can be configured, executed, and analyzed within Flare. We have paid special attention to the key symptoms to watch out for when assessing the stability, and thus accuracy, of your membrane protein MD simulations.
For a visual demonstration of Flare's molecular dynamics environment and workflows and analysis tools, watch our webinar.